Introduction & strategy
Many therapeutics are nowadays produced in cells. When biopharmaceutical companies develop new Biologics, they usually go through different steps including Bioprocess optimization, with the ultimate goal to increase production yield and purity of the drug product. However, getting a better drug substance requires production and/or purification processes modifications, potentially modifying product purity or efficiency. Consequently, quality control analysis and biological activity assessment are required, particularly batch comparison for Critical Quality Attributes (CQA), including Host Cell Protein analysis (HCP analysis). In this study, we investigated why a Biologic excreted in the extracellular media lacks its catalytic activity in 1 out of 3 production batches, consequently to process optimization.
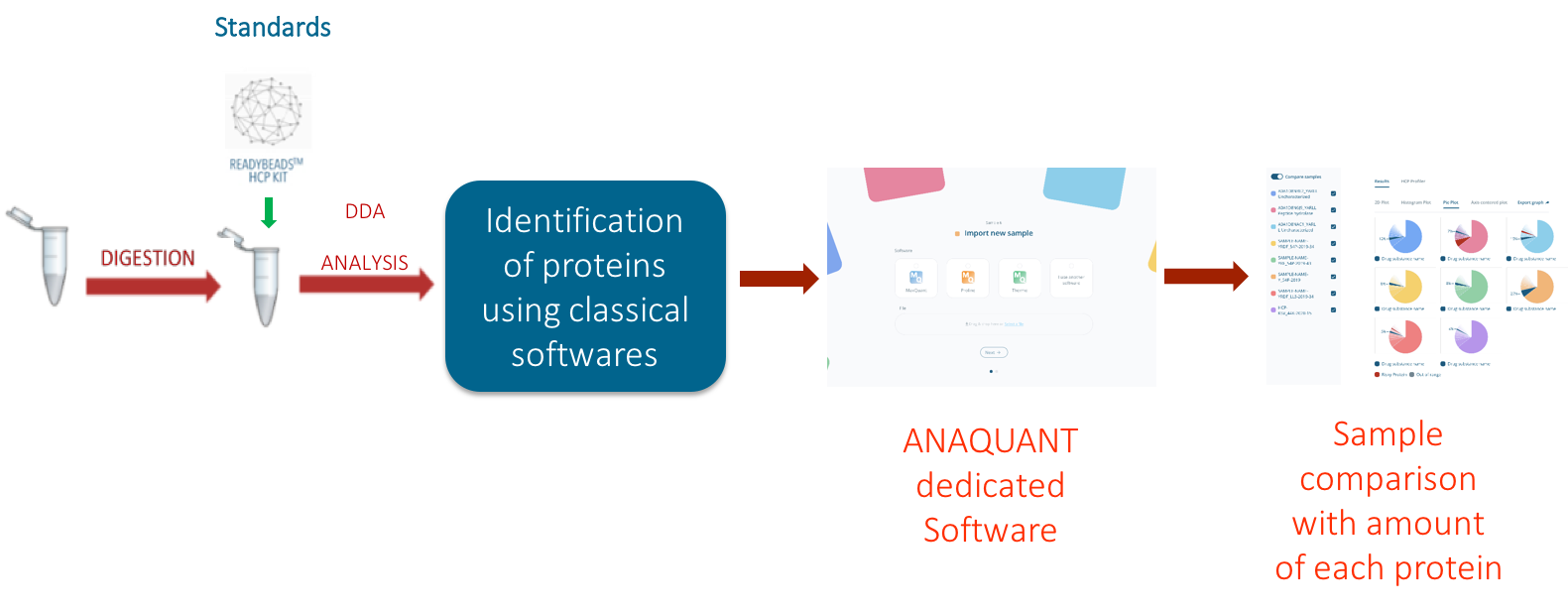
Figure 1: HCP PROFILER integrated workflow for Host cell protein analysis
We deployed a MS-based assay called HCP-PROFILER (figure 1), to individually identify and quantify each HCP in a single MS analysis. This solution is based on an innovative standard relying on a hydro-soluble bead containing an internal calibration curve including several peptides at well-defined concentration levels; and integrated software for user-friendly data management and viewing. The standard is added into the sample before MS analysis, and calibration curve allows overpassing instrumental variations, to compare batches produced over months. To the extent of our knowledge, this approach brings a unique analytical solution, compared to classical approaches, such as Enzyme-linked immunosorbent assay (ELISA), which suffer from several limits. Indeed, ELISA is commonly used to monitor global HCPs levels. Nevertheless, it does not allow comprehensive HCP identification and only provides total HCP amount, most often in arbitrary units.
Results
After identification step, according to a database search strategy, individual HCPs were identified in each batch. Results demonstrated that compared to other batches, the third one owns 3-fold more HCPs (figure 2). To explain the so-observed differences, more investigations were required. It was especially observed that a large part of proteins didn’t own signal peptide necessary for protein excretion in extracellular media. Thus, compared to batches 1 & 2, it was possible to suggest probable cell lysis consequently to cell death, induced by increased incubation time in the optimized process.
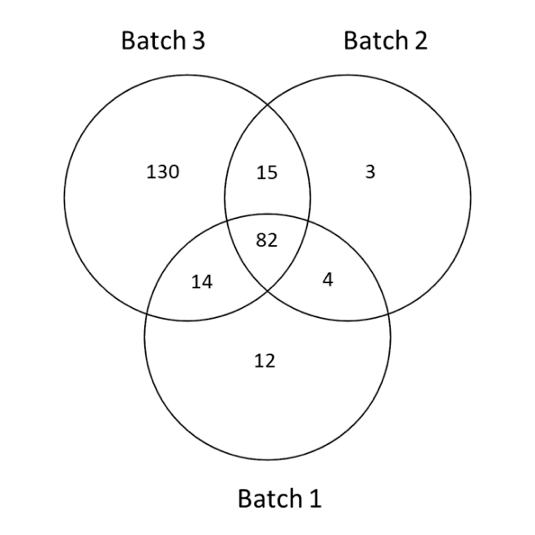
Figure 2: Venn-plot obtained from the comparison of identified host cell proteins in 3 batches
The real input of HCP-PROFILER integrated workflow consists of both, identify and assess individual accurate quantities of HCPs in a single MS analysis. Moreover, according to it normalization capabilities HCP-PROFILER allows batches comparison produced over months, at a quantitative level. Thus, comparison of quantification results was done for the 3 batches. Results demonstrated that among the 130 HCPs specifics to batch 3, 5 were found with significant quantity difference (up to 4-fold) compared to the 2 others batches, while 2 were only found in batch 3.
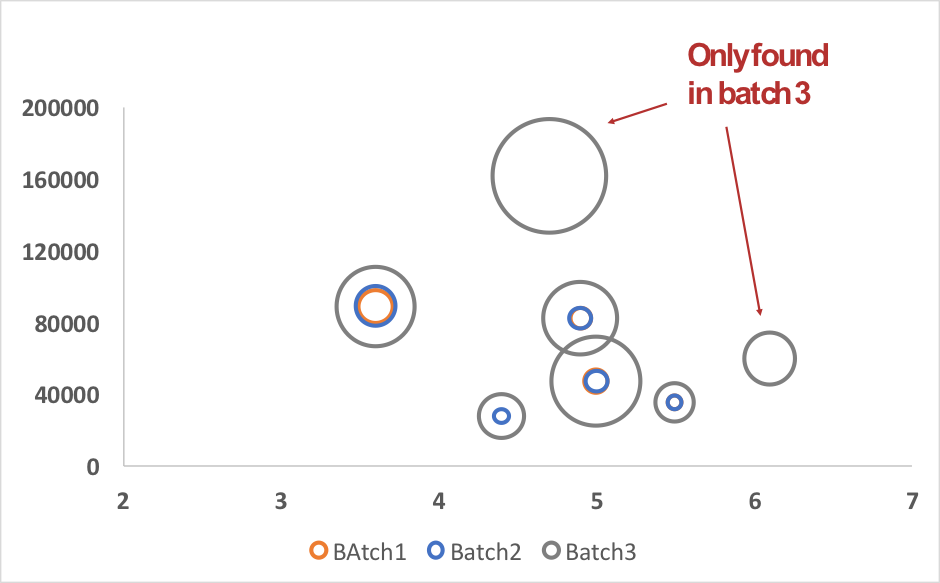
Figure 3: 2D-plot obtained for 7 protein over-expressed in batch 3. In this plot, circles represent individual identified proteins and size of each circle represent amount of protein.
Among the 7 differentially expressed proteins, two proteins particularly draw the attention due to their proteolytic activities. These MS-based results raised judicious ways to investigate, to try to explain the observed lack of activity.
Discussion / Conclusion
The objective of this study was to investigate why after process optimisation, a biologic didn’t bring any catalytic activity. So, critical quality attribute monitoring, particularly HCPs, appeared a judicious way to explore, to assess whether risky HCPs could be incriminated. Since classical approaches such as ELISA can not extract individual identity, we developed HCP-PROFILER integrated workflow. Results demonstrated the power of this approach to individually identify and quantify HCPs in a single MS-based analysis, with an easy and straightforward pipeline. Moreover, this approach gave precious information to understand the observed differences. First, identified proteins lead to the understanding that cell lysis probably occurred during the Upstream Process (USP). Second, quantitative results demonstrated some proteins as significantly over-expressed. Particularly risky HCPs were highlighted, owning hydrolase inhibitor activity or proteolytic activity. Moreover, third batch comparison over months is ensured to be possible thanks to the universal internal standards properties bring by HCP-PROFILER and the READYBEADS technology. To conclude, these crucial pieces of informations indicate the way to follow to push forward optimization process development.
 Anaquant HCP analysis I Protein characterisation I Protein analysis
Anaquant HCP analysis I Protein characterisation I Protein analysis