Introduction
Therapeutic biologics purification steps are developed to remove impurities and host cell proteins (HCP). Residual proteins monitoring or HCP analysis are strongly encouraged by regulatory, since they can affect product quality, stability, efficacy and safety, or induce immunogenicity. From the cells harvest to final drug substance (DS), several purification steps are implemented and optimized through a Down Stream Process (DSP). However, DSP optimization can affect HCP profile, and it is mandatory to assess HCP identification after each optimization development. In this study, we investigate HCPs profile modifications related to DSP optimization tests. Indeed, following the production and the analysis of a reference batch, a new batch was produced with new DSP parameters. After quality control analysis based on LC-UV, two new contaminants were detected. It didn’t appear very clear whether it was contaminants or degradation products. Thus, a specific and reproducible approach was necessary to answer to this issue. HCP-PROFILER integrated workflow appeared to be the best appropriate solution, in regards to it capabilities for simultaneous identification and quantification of HCP in a single MS analysis, with an easy-to-use and integrated analytical workflow.
Results
We deployed HCP-PROFILER integrated workflow to explore the HCP profile in 3 DS batches (HCP identification and quantification). HCP-PROFILER is composed of an innovative standard relying on a hydro-soluble bead containing an internal calibration curve including several peptides at well-defined concentration levels and a dedicated software to convert MS signal into amount of protein. The standard is spiked directly into sample and solves, thanks to the READYBEADS technology, reproducibility concerns over time with standard stability ensured over several months. Also, according to its capabilities to normalize instrumental or experimental-induced variations, this new workflow allows DS batch comparison over several months and thus provides a technical solution for easy and robust batch-to-batch comparison or downstream process performance assessment (figure 1). That is why this workflow represented the best analytical strategy to answer to the study purpose.
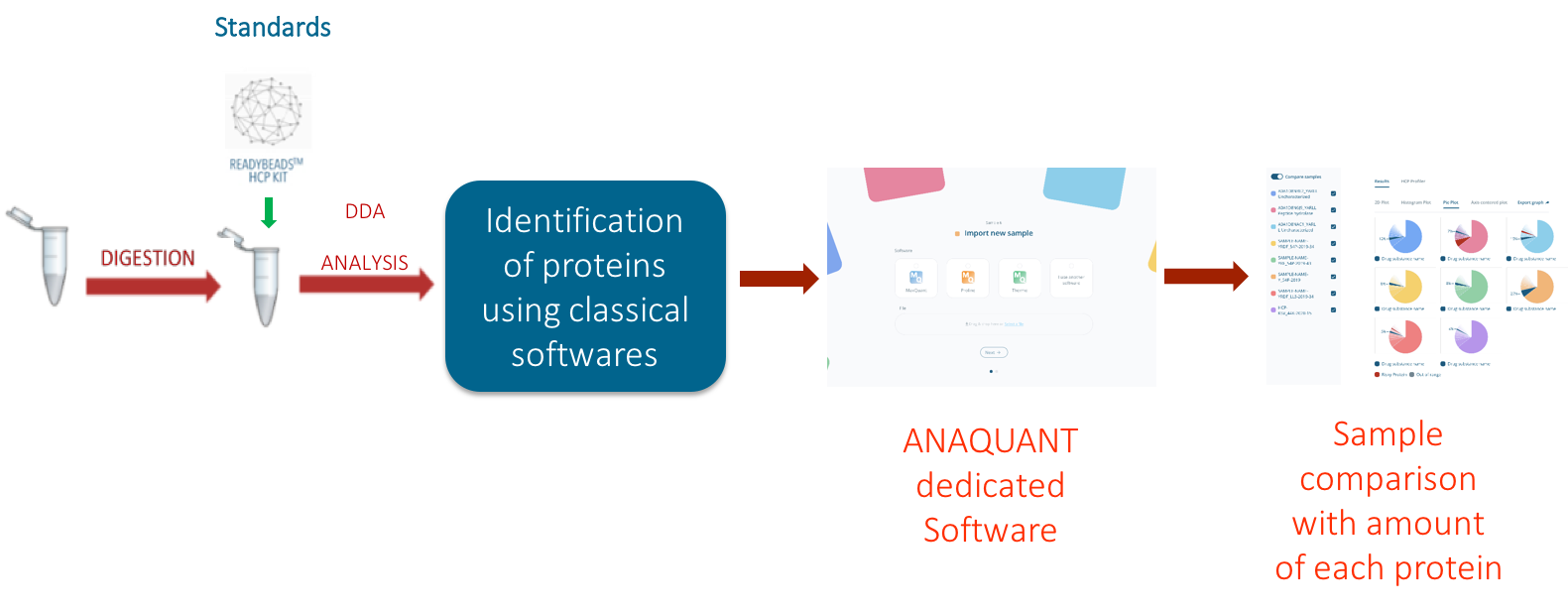
Figure 1: Schematic representation of HCP-PROFILER integrated workflow
To improve data consistency, a QC protein was added in each sample to verify the robustness and accuracy of the analytical workflow. Then, after tryptic digestion, 1 READYBEADS™ of HCP-PROFILER standard was added in each sample before to be injected for LC MS analysis. Data processing was performed and integrated software allowed user-friendly data management and comparison. Results in figure 2 indicates, for batch 1, 27 quantified proteins in a range from 16 to 11 068 ppm (protein 1 found at 11 068 ppm).
In batch 2, corresponding to a modified DSP developed to eliminate the major impurity, it surprisingly appeared that 41 proteins were quantified in a range from 4 to 21 100 ppm. 2 contaminants were clearly identified as Host Cell Protein, and did not correlate with a degradation of the product. These proteins (Protein 2 & Protein 3 on figure 2) were respectively quantified at 21 100 and 12 000 ppm, whereas Protein 1 was found at 1 186 ppm. Clearly, these results were of great interest for both characterizing newly observed contaminants and quantifying these latter. Concentration results concluded that observed levels were incompatible with regulatory guidelines.
Finally, batch 3 resulting from the latest DSP generation was analysed. Results demonstrated 25 quantified proteins, in a range from 10 to 2 150 ppm. Protein 2 and Protein 3, were found at 74 and 25 ppm, respectively. Protein 1 was found at 52 ppm.
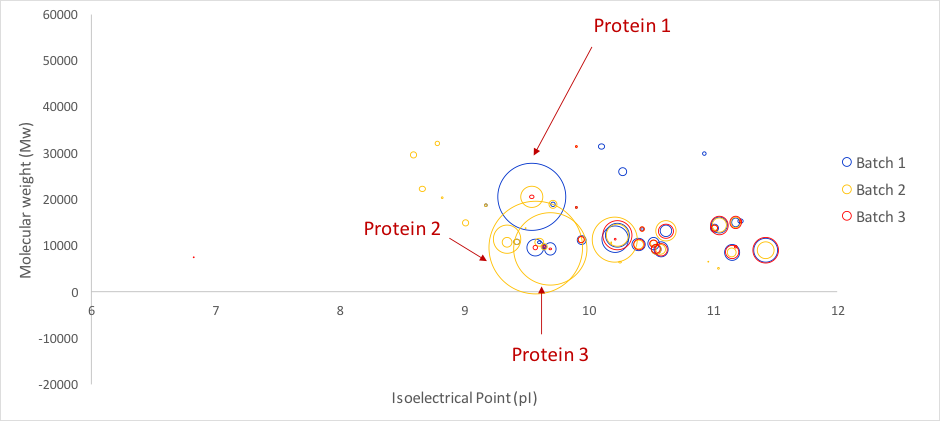
Figure 2: Comparison of HCPs profiles for 3 DS batches with HCP-PROFILER integrated workflow
Conclusion
This study demonstrated that HCP-PROFILER integrated workflow brings a new way to compare DS batches through times. Indeed, it was possible to identify 2 previously observed but unknown contaminants, to simultaneously quantify their amounts and also to evaluate DSP optimization through development steps over months. In this study, the comparison allowed observation of a huge decrease of HCPs through DSP optimization steps. Information generated thanks to the HCP profiler solution could be of great help to improve Downstream process steps, improve drug development and be in accordance with the recommendations of the authorities.
 Anaquant HCP analysis I Protein characterisation I Protein analysis
Anaquant HCP analysis I Protein characterisation I Protein analysis