Introduction
mRNA-based cancer therapies and vaccines against infectious diseases are becoming increasingly prominent as new therapeutics. These therapeutics can be manufactured using In Vitro Transcription platforms. However, the safety and efficiency of mRNA therapeutics may be impacted by impurities generated during the production process.
During biological development, manufacturers must demonstrate the purity of the drug and the consistency of the manufacturing process in relation to potential contaminants. These contaminants are most often process-related protein impurities such as RNA polymerase, restriction enzymes, or DNase, frequently used during the manufacture of RNA drugs. Additionally, Host Cell Proteins (HCP) from the production system can pose significant challenges.
To address these issues, ANAQUANT has developed HCPprofiler, an innovative solution dedicated to MS-based impurity profiling and Host Cell Protein analysis. This solution is composed by both
A hydro-soluble bead coated with peptides based on the READYBEADS technology, which, after being spiked directly into the sample, releases peptides at well-controlled concentrations. This enables the generation of a robust and universal calibration curve.
A dedicated software that converts MS signals into quantitative data for each identified protein. The selected peptides are non-natural, with sequences that do not correspond to any proteome available in public databases. As a result, these peptides can be spiked into all samples regardless of taxonomy, without generating interference.
In this application note we performed process related proteins and HCPs identification and quantification inside two different batches of mRNA therapeutic drug at two different stages of the downstream process, a middle step and the final drug substance step.
Method
In the strategy implemented, all proteins present in the samples are digested enzymatically. After that, cation exchange chromatography was implemented to separate the RNA from the peptide digest before LC-MS/MS analysis in DDA (Data Dependent Analysis) mode.
Results
LC-MS analysis of the four different mRNA samples, mid DownStream Process (DSP) samples and drug substances (DS) allowed to identify a small number of process related impurities. Batch consistency was shown with only two process related impurities (one known protein added during the process and one HCP) in both DS batches.
HCPprofiler internal standards added to each four-sample preparations allowed to obtain universal calibration curves used both for sample injection normalisation and individual protein quantification.
Three process-related impurities and 3 HCPs are quantified in the two intermediate DSP step samples (Figure 2). The identification of these proteins and information on their physicochemical properties are important and will make it possible to improve the DSP process.
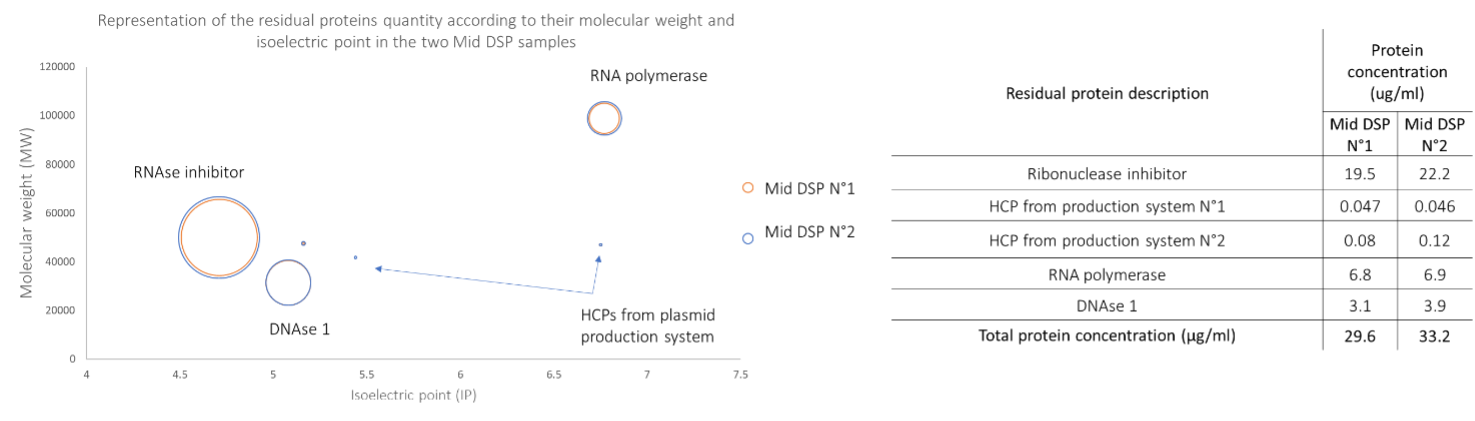
Figure 2 : Left, 2D-plot graphic that represents the calculated protein quantity depending on its molecular weight and isoelectric point. Right, calculated process related impurities concentration in both middle downstream process samples
Individual HCPs quantification was also performed allowing to go deeper into the DS samples characterisation: enabling estimation of process related protein impurities abundances in ng/ml or ppm (see Figure 3) in both DS samples.
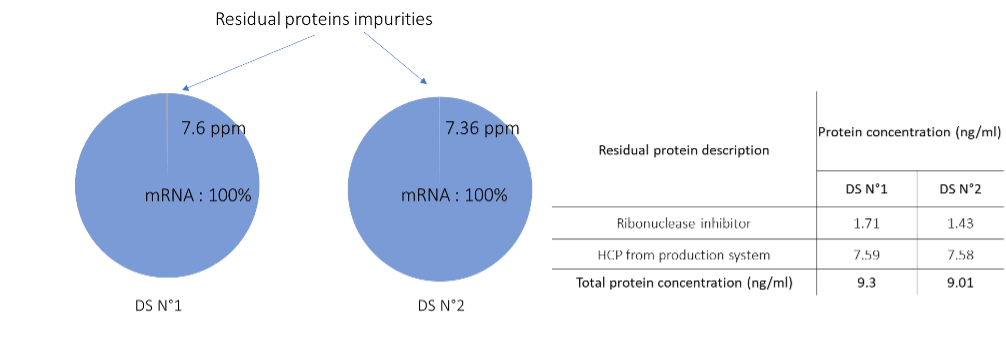
Figure 3 : Global process related protein impurities quantification results. Left, pie plots of the mRNA purity in each Drug Substance sample. Right, individual protein quantifications
The overall mRNA purity as well as the individual residual proteins quantities are shown for both mRNA DS using the HCPprofiler solution. Regarding the results obtained in both mid step DSP samples and in DS samples it clearly appears that the actual downstream process allows the known related process impurities purification that decreases from 20µg/ml to almost 1.5ng/ml for the ribonuclease inhibitor protein. On the contrary, unknown HCPs only decrease from almost 50ng/ml to almost 7.6 ng/ml. This highlights the importance of knowing the impurities to adapt the purification process.
Conclusion
The analytical workflow presented enables the highly sensitive identification of process-related impurities, including both known added enzymes and Host Cell Proteins (HCPs), in mRNA therapeutic samples across different stages of the downstream processing (DSP). With the HCPprofiler solution, not only does it provide precise protein identification, but it also allows for individual quantification of residual protein impurities in µg/ml.
This deeper level of sample characterization enables the identification of potentially immunogenic HCPs from the production system. The accurate identification of these HCPs is critical, as it can inform and optimize the purification process to enhance the overall safety and efficacy of the therapeutic product.
By leveraging HCPprofiler, manufacturers can gain actionable insights into their process impurities, ultimately leading to a more refined, safe, and consistent production pipeline.
 Anaquant HCP analysis I Protein characterisation I Protein analysis
Anaquant HCP analysis I Protein characterisation I Protein analysis