
Introduction
Modified animal models, specifically humanized mouse models, are widely used in biological and biotechnological research. These models are obtained through gene sequence modifications to alter, modify, or induce protein expression. This approach, implemented to induce human protein expression in animal models or cells, is particularly used to evaluate drug efficacy on the targeted human protein.
In order to ensure model validity, it is mandatory to verify that the genetic modifications are accurately reflected in protein expression. However, the expressed human protein often has high sequence homology with the corresponding animal protein. Thus, immunological tests often lack the specificity required, due to the similarity in sequences, making them difficult to use.
LC-MS (Liquid Chromatography-Mass Spectrometry) is currently considered an orthogonal approach that, due to its specificity properties, can unambiguously identify and quantify proteins with high sequence homologies. In this note, we demonstrated the added value of the LC-SRM (Liquid Chromatography-Selected Reaction Monitoring) approach to monitor human protein expression in humanized mouse models.
Results
In this study, expressed human protein was monitored in humanized mouse models. These models are crucial in preclinical studies, especially for oncology, immuno-oncology, and infectious disease research. “Signature peptides” specific to the human protein region were only present in modified cell lines, whereas shared peptides were present in both the wild-type (WT) and modified models. Immuno-based assays were not specific enough to discriminate proteins with a high degree of homology in their sequences, making LC-SRM the best strategy due to its specificity and sensitivity. Three signature peptides were selected based on their specificity for detecting the protein of interest. Proteins extracted from cells and tissues of both Human+ and WT models were enzymatically digested. The resulting peptide mix was analyzed using LC-SRM. Results demonstrated that human protein signature peptides were only detected in Human+ models. Shared peptides corresponding to endogenous mouse protein peptides were found in both Human+ and WT models, confirming the specificity of LC-SRM in identifying human proteins within a humanized mouse model (Figure 1).
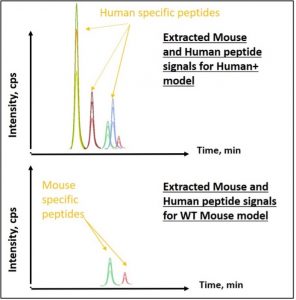
Figure 1 : Chromatogram of extracted peptides from mouse or human protein. Top) Human + model expressing the Human protein in addition to the mouse endogenous protein; bottom) WT model expressing only the mouse protein.
These results validated the Human+ models and confirmed the power of LC-SRM (Liquid Chromatography-Selected Reaction Monitoring) to discriminate and monitor human protein expression in humanized mouse models. This analytical approach ensures a quick and unambiguous response regarding the validity of Human+ models. This method was used to test two modified Human+ cell models, along with two control (WT) models, as previously described. Results demonstrated that human protein was expressed in both Human+ models (Figure 2), while it was not detected in the WT models.
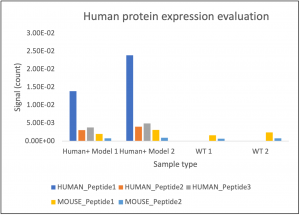
Figure 2 : Graphic representing the HUMAN or MOUSE specific peptide signal for different cell models (WT; and Human+ models)
Concluding remarks
In this study, the LC-SRM approach was developed to validate Human+ models. Due to its specificity properties, it was the only valuable approach to discriminate two proteins belonging to different taxonomies but with a high degree of homology. Based on this quick analytical approach, two Human+ models were validated. Usually, such Knock-in models generate a high protein expression, enabling the unambiguous observation of human protein-specific peptides, with a high LC-MS/MS signal intensity.
 Anaquant HCP analysis I Protein characterisation I Protein analysis
Anaquant HCP analysis I Protein characterisation I Protein analysis