Introduction
Knockout cell or animal model are widely used in biological/biotechnological researches, and consists in silencing protein function, either by gene deletion or gene sequence modification to alter protein function. This approach is widely used to elucidate gene function, metabolic disorders induced by specific gene dysfunction, or to obtain strains free of risky proteases. In order to control KO model validity, it is mandatory to control whether the genetic modifications are reflected after protein expression. However, the resulting expressed product can be chimera proteins, truncated proteins, or protein with modified sequences. Consequently, the use of immunological tests appears to be difficult in regards of the lake of antibody specificity. LC-MS is currently considered as an orthogonal approach, which, thanks to it specificity properties, is able to unambiguously identify and quantify proteins with really close sequences. In this note, we demonstrated the input of LC-SRM approach to validate a KO model.
Results
In this study, expressed protein in KO model is a truncated one, consequently, “signature peptides” are only present in wild type cell line, while they are absent in the KO model. Immuno-based assays were not enough specific to discriminate so close protein sequences. Thus, LC SRM appeared the best strategy. Two signature peptides were selected, based on their specificity to detect protein of interest. Then, extracted proteins from cells/tissues were enzymatically digested, for KO and WT models. Consequently, peptide mix was obtained and analysed by LC-SRM. Results demonstrated that signature peptides were only detected in WT models, while shared peptides were found at equal intensity in both models (Figure 1).
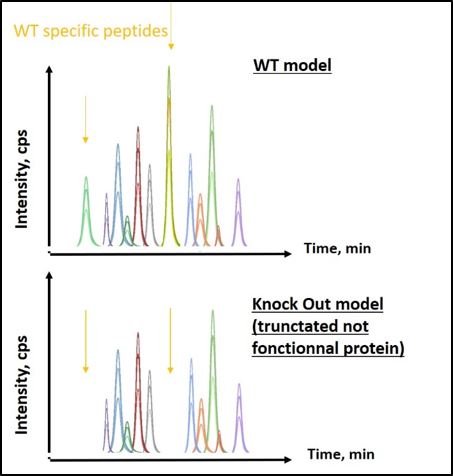
Figure 1 : Chromatogram of extracted peptides from protein 1. Top) WT model expressing the entire functional protein; bottom) KO model expressing a truncated not functional protein
These results allowed validation of KO models, and confirmed the power of LC-SRM to discriminate KO from WT models. This kind of rapid analysis ensures a rapid and unambiguous response about KO models validity. This was used to test 3 supplemental KO models (one in duplicate) after genetic modifications. For that, 3 controls (WT models) and 3 KO models were analyzed as previously described. Results demonstrated that 1 out of 3 models was not KO since signature peptides were detected (Figure 2).
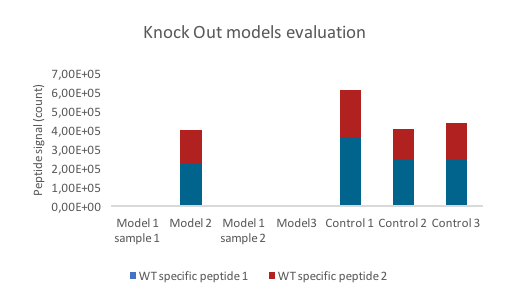
Figure 2 : Graphic representing the WT specific peptide signal for different cell models (control =WT; other models = expected KO models)
Concluding remarks
In this study, LC-SRM approach was developed to validate KO models. According to its specificity, it was the only valuable approach to discriminate KO truncated proteins from WT protein. Based on this rapid analytical approach, 1 KO model was identified as invalid, which is a key information before developing further experiment.
 Anaquant HCP analysis I Protein characterisation I Protein analysis
Anaquant HCP analysis I Protein characterisation I Protein analysis